We use text mining a lot in day-to-day data mining operations. In order to share our knowledge on this, to show that R is an extremely mature platform to do business-oriented text analytics and to give you practical experience with text mining, our course on Text Mining with R is scheduled for the 3rd consecutive year at LStat, the Leuven Statistics Research Center (Belgium) as well as at the Data Science Academy in Brussels. Courses are scheduled 2 times in November 2017 and also in March 2018.
 This course is a hands-on course covering the use of text mining tools for the purpose of data analysis. It covers basic text handling, natural language engineering and statistical modelling on top of textual data. The following items are covered.
This course is a hands-on course covering the use of text mining tools for the purpose of data analysis. It covers basic text handling, natural language engineering and statistical modelling on top of textual data. The following items are covered.
- Text encodings
- Cleaning of text data, regular expressions
- String distances
- Graphical displays of text data
- Natural language processing: stemming, parts-of-speech tagging, tokenization, lemmatisation
- Sentiment analysis
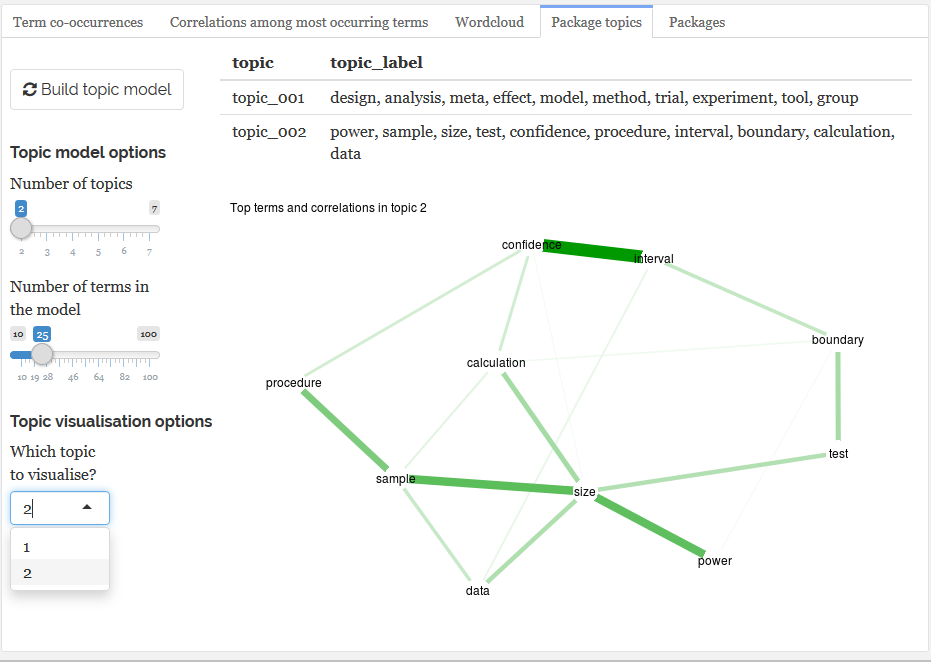
- Statistical topic detection modelling and visualization (latent diriclet allocation)
- Visualisation of correlations & topics
- Word embeddings
- Document similarities & Text alignment
Feel free to register at the following dates:
- 18-19/10/2017: Statistical machine learning with R. Leuven (Belgium). Subscribe here
- 08+10/11/2017: Text mining with R. Leuven (Belgium). Subscribe here
- 27-28/11/2017: Text mining with R. Brussels (Belgium). http://di-academy.com/bootcamp + send mail to This email address is being protected from spambots. You need JavaScript enabled to view it.
- 19-20/12/2017: Applied spatial modelling with R. Leuven (Belgium). Subscribe here
- 20-21/02/2018: Advanced R programming. Leuven (Belgium). Subscribe here
- 08-09/03/2018: Computer Vision with R and Python. Leuven (Belgium). Subscribe here
- 22-23/03/2018: Text Mining with R. Leuven (Belgium). Subscribe here
If you work on natural language processing in a day-to-day setting which involves statistical engineering, at a certain timepoint you need to process your text with a number of text mining procedures of which the following ones are steps you must do before you can get usefull information about your text
- Tokenisation (splitting your full text in words/terms)
- Parts of Speech (POS) tagging (assigning each word a syntactical tag like is the word a verb/noun/adverb/number/...)
- Lemmatisation (a lemma means that the term we "are" is replaced by the verb to "be", more information: https://en.wikipedia.org/wiki/Lemmatisation)
- Dependency Parsing (finding relationships between, namely between "head" words and words which modify those heads, allowing you to look to words which are maybe far away from each other in the raw text but influence each other)
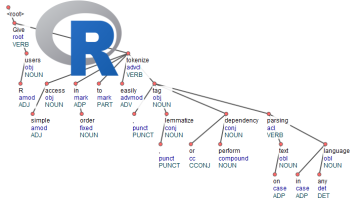
If you do this in R, there aren't much available tools to do this. In fact there are none which
- do this for multiple language
- do not depend on external software dependencies (java/python)
- which also allow you to train your own parsing & tagging models.
Except R package udpipe (https://github.com/bnosac/udpipe, https://CRAN.R-project.org/package=udpipe) which satisfies these 3 criteria.
If you are interested in doing the annotation, pre-trained models are available for 50 languages (see ?udpipe_download_model) for details. Let's show how this works on some Dutch text and what you get of of this.
library(udpipe)
dl <- udpipe_download_model(language = "dutch")
dl
language file_model
dutch C:/Users/Jan/Dropbox/Work/RForgeBNOSAC/BNOSAC/udpipe/dutch-ud-2.0-170801.udpipe
udmodel_dutch <- udpipe_load_model(file = "dutch-ud-2.0-170801.udpipe")
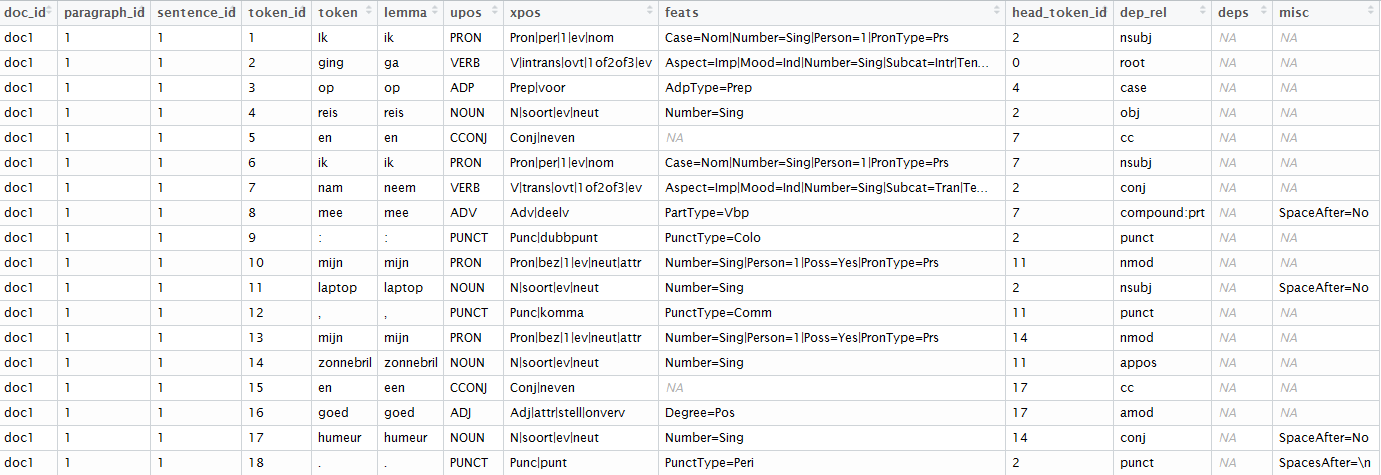
x <- udpipe_annotate(udmodel_dutch,
x = "Ik ging op reis en ik nam mee: mijn laptop, mijn zonnebril en goed humeur.")
x <- as.data.frame(x)
x
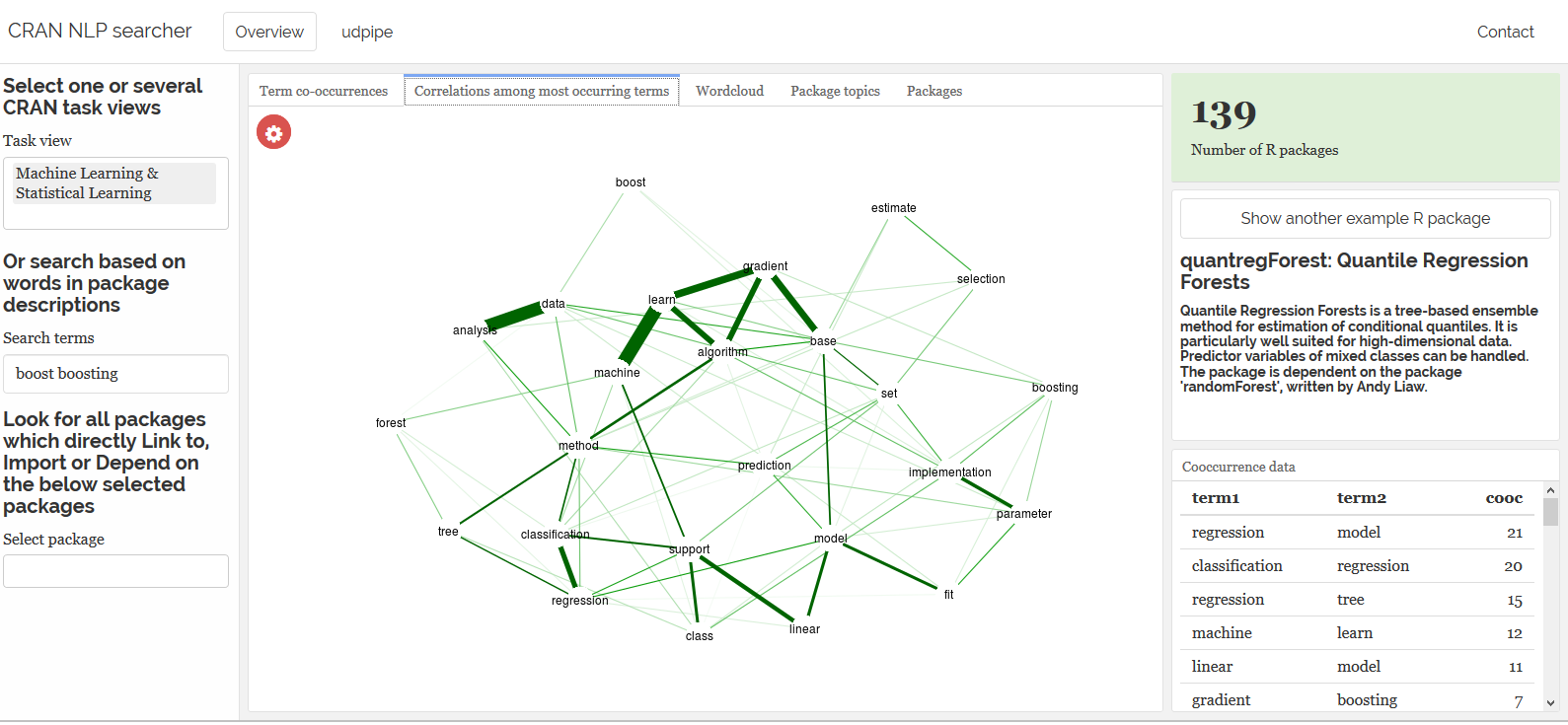
The result of this is a dataset where text has been splitted in paragraphs, sentences, words, words are replaced by their lemma (ging > ga, nam > neem), you get the universal parts of speech tags, detailed parts of speech tags, you get features of the word and with the head_token_id we see which words are influencing other words in the text as well as the dependency relationship between these words.
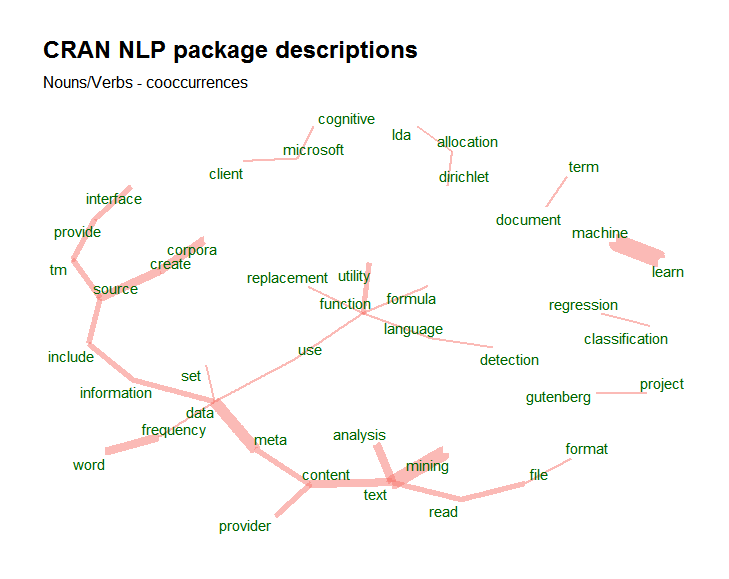
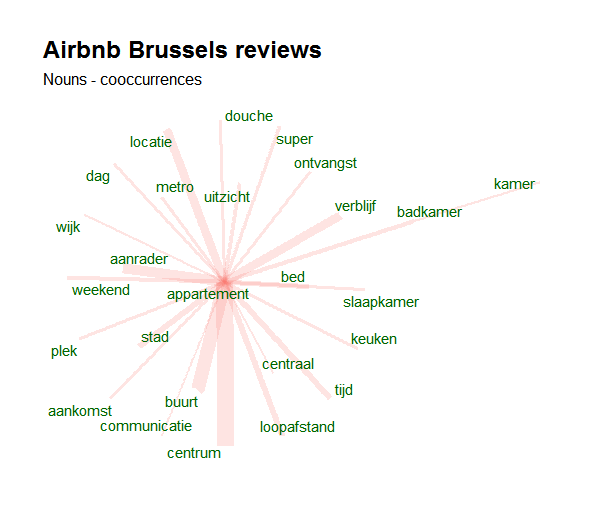
To go from that dataset to meaningfull visualisations like this one is than just a matter of a few lines of code. The following visualisation shows the co-occurrence of nouns with customer feedback on Airbnb appartment stays in Brussels (open data available at http://insideairbnb.com/get-the-data.html).
In a next post, we'll show how to train your own tagging models.
If you like this type of analysis or if you are interested in text mining with R, we have 3 upcoming courses planned on text mining. Feel free to register at the following links.
- 18-19/10/2017: Statistical machine learning with R. Leuven (Belgium). Subscribe here
- 08+10/11/2017: Text mining with R. Leuven (Belgium). Subscribe here
- 27-28/11/2017: Text mining with R. Brussels (Belgium). http://di-academy.com/bootcamp + send mail to This email address is being protected from spambots. You need JavaScript enabled to view it.
- 19-20/12/2017: Applied spatial modelling with R. Leuven (Belgium). Subscribe here
- 20-21/02/2018: Advanced R programming. Leuven (Belgium). Subscribe here
- 08-09/03/2018: Computer Vision with R and Python. Leuven (Belgium). Subscribe here
- 22-23/03/2018: Text Mining with R. Leuven (Belgium). Subscribe here
For business questions on text mining, feel free to contact BNOSAC by sending us a mail here.
For R users interested in Machine Learning, you can attend our upcoming course on Machine Learning with R which is scheduled on 18-19 October 2017 in Leuven, Belgium. This is now the 4th year this course is given at the university of Leuven so we made quite some updates since the first time this was given 4 years ago.
During the course you'll learn the following techniques from a methodological as well as a practical perspective: naive bayes, trees, feed-forward neural networks, penalised regression, bagging, random forests, boosting and if time permits graphical lasso, penalised generalised additive models, support vector machines.
Subscribe here: https://lstat.kuleuven.be/training/coursedescriptions/statistical-machine-learning-with-r

For a full list of training courses provided by BNOSAC - either in-house or in-public: go to http://www.bnosac.be/training
For R users interested in text mining with R, applied spatial modelling with R, advanced R programming or computer vision, you can also subscribe for the following courses, scheduled at the University of Leuven.
- 18-19/10/2017: Statistical machine learning with R. Leuven (Belgium). Subscribe here
- 08+10/11/2017: Text mining with R. Leuven (Belgium). Subscribe here
- 19-20/12/2017: Applied spatial modelling with R. Leuven (Belgium). Subscribe here
- 20-21/02/2018: Advanced R programming. Leuven (Belgium). Subscribe here
- 08-09/03/2018: Computer Vision with R and Python. Leuven (Belgium). Subscribe here
- 22-23/03/2018: Text Mining with R. Leuven (Belgium). Subscribe here
Just before the summer holidays, BNOSAC presented a talk called Computer Vision and Image Recognition algorithms for R users at the UseR conference. In the talk 6 packages on Computer Vision with R were introduced in front of an audience of about 250 persons. The R packages we covered and that were developed by BNOSAC are:
- image.CornerDetectionF9: FAST-9 corner detection
- image.CannyEdges: Canny Edge Detector
- image.LineSegmentDetector: Line Segment Detector (LSD)
- image.ContourDetector: Unsupervised Smooth Contour Line Detection
- image.dlib: Speeded up robust features (SURF) and histogram of oriented gradients (FHOG) features
- image.darknet: Image classification using darknet with deep learning models AlexNet, Darknet, VGG-16, GoogleNet and Darknet19. As well object detection using the state-of-the art YOLO detection system
For those of you who missed this, you can still see the video of the presentation & view the pdf of the presentation below. The packages are open-sourced and made available at https://github.com/bnosac/image
If you have a computer vision endaveour in mind, feel free to get in touch for a quick chat. For those of you interested in following training on how to do image analysis, you can always register for our training on Computer Vision with R and Python here. More details on the full training program and training dates provided by BNOSAC: visit http://bnosac.be/index.php/training
 This course is a hands-on course covering the use of text mining tools for the purpose of data analysis. It covers basic text handling, natural language engineering and statistical modelling on top of textual data. The following items are covered.
This course is a hands-on course covering the use of text mining tools for the purpose of data analysis. It covers basic text handling, natural language engineering and statistical modelling on top of textual data. The following items are covered.



