bigglm on your big data set in open source R, it just works - similar as in SAS
In a recent post by Revolution Analytics (link & link) in which Revolution was benchmarking their closed source generalized linear model approach with SAS, Hadoop and open source R, they seemed to be pointing out that there is no 'easy' R open source solution which exists for building a poisson regression model on large datasets.

- Download the data
require(ffbase)
require(LaF)
require(ETLUtils)
download.file("http://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/BSAPUFS/Downloads/2010_Carrier_PUF.zip", "2010_Carrier_PUF.zip")
unzip(zipfile="2010_Carrier_PUF.zip")
- Import it (showing 2 options - either by using package LaF or with read.csv.ffdf using argument transFUN to recode the input data according to the codebook which you can find here)
## the LaF package is great if you are working with fixed-width format files but equally good for csv files and laf_to_ffdf does what it ## has to do: get the data in an ffdf dat <- laf_open_csv(filename = "2010_BSA_Carrier_PUF.csv",
column_types = c("integer", "integer", "categorical", "categorical", "categorical", "integer", "integer", "categorical", "integer", "integer", "integer"), column_names = c("sex", "age", "diagnose", "healthcare.procedure", "typeofservice", "service.count", "provider.type", "servicesprocessed", "place.served", "payment", "carrierline.count"), skip = 1) x <- laf_to_ffdf(laf = dat) ## the transFUN is easy to use if you want to transform your input data before putting it into the ffdf, ## it applies a function to your read input data which is read in in chunks ## We use it here to recode the numbers to factors according to the code book which you can find in the codebook x <- read.csv.ffdf(file = "2010_BSA_Carrier_PUF.csv", colClasses = c("integer","integer","factor","factor","factor","integer","integer","factor","integer","integer","integer"), transFUN=function(x){ names(x) <- recoder(names(x), from = c("BENE_SEX_IDENT_CD", "BENE_AGE_CAT_CD", "CAR_LINE_ICD9_DGNS_CD", "CAR_LINE_HCPCS_CD", "CAR_LINE_BETOS_CD", "CAR_LINE_SRVC_CNT", "CAR_LINE_PRVDR_TYPE_CD", "CAR_LINE_CMS_TYPE_SRVC_CD", "CAR_LINE_PLACE_OF_SRVC_CD", "CAR_HCPS_PMT_AMT", "CAR_LINE_CNT"), to = c("sex", "age", "diagnose", "healthcare.procedure", "typeofservice", "service.count", "provider.type", "servicesprocessed", "place.served", "payment", "carrierline.count")) x$sex <- factor(recoder(x$sex, from = c(1,2), to=c("Male","Female"))) x$age <- factor(recoder(x$age, from = c(1,2), to=c("Under 65", "65-69", "70-74", "75-79", "80-84", "85 and older"))) x$place.served <- factor(recoder(x$place.served, from = c(0, 1, 11, 12, 21, 22, 23, 24, 31, 32, 33, 34, 41, 42, 50, 51, 52, 53, 54, 56, 60, 61, 62, 65, 71, 72, 81, 99), to = c("Invalid Place of Service Code", "Office (pre 1992)", "Office","Home","Inpatient hospital","Outpatient hospital", "Emergency room - hospital","Ambulatory surgical center","Skilled nursing facility", "Nursing facility","Custodial care facility","Hospice","Ambulance - land","Ambulance - air or water", "Federally qualified health centers", "Inpatient psychiatrice facility", "Psychiatric facility partial hospitalization", "Community mental health center", "Intermediate care facility/mentally retarded", "Psychiatric residential treatment center", "Mass immunizations center", "Comprehensive inpatient rehabilitation facility", "End stage renal disease treatment facility", "State or local public health clinic","Independent laboratory", "Other unlisted facility"))) x }, VERBOSE=TRUE) class(x) dim(x)
- Profile your data
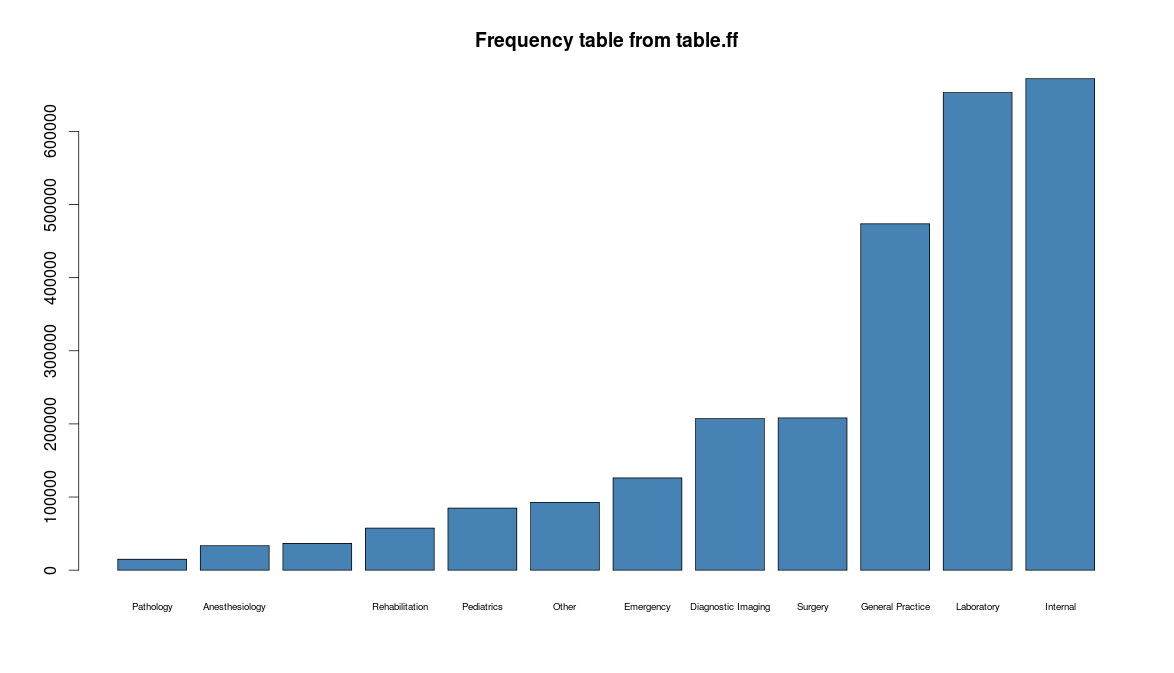
## ## Data Profiling using table.ff ## table.ff(x$age) table.ff(x$sex) table.ff(x$typeofservice) barplot(table.ff(x$age), col = "lightblue") barplot(table.ff(x$sex), col = "lightblue") barplot(table.ff(x$typeofservice), col = "lightblue")
- Grouping by - showing the speedy binned_sum
##
## Basic & fast group by with ff data
##
doby <- list()
doby$sex <- binned_sum.ff(x = x$payment, bin = x$sex, nbins = length(levels(x$sex)))
doby$age <- binned_sum.ff(x = x$payment, bin = x$age, nbins = length(levels(x$age)))
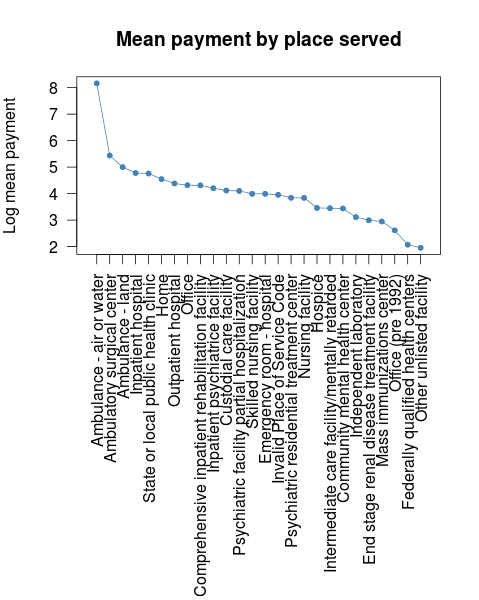
doby$place.served <- binned_sum.ff(x = x$payment, bin = x$place.served, nbins = length(levels(x$place.served)))
doby <- lapply(doby, FUN=function(x){
x <- as.data.frame(x)
x$mean <- x$sum / x$count
x
})
doby$sex$sex <- recoder(rownames(doby$sex), from = rownames(doby$sex), to = levels(x$sex))
doby$age$age <- recoder(rownames(doby$age), from = rownames(doby$age), to = levels(x$age))
doby$place.served$place.served <- recoder(rownames(doby$place.served), from = rownames(doby$place.served), to = levels(x$place.served))
doby
- Build a generalized linear model using package biglm which integrates with ffbase::bigglm.ffdf
##
## Make a linear model using biglm
##
require(biglm)
mymodel <- bigglm(payment ~ sex + age + place.served, data = x)
summary(mymodel)
# This will overflow your RAM as it will get your data from ff into RAM
#summary(glm(payment ~ sex + age + place.served, data = x[,c("payment","sex","age","place.served")]))
- Do the same on more data: 280Mio records
##
## Ok, we were working only on +/- 2.8Mio records which is not big, let's explode the data by 100 to get 280Mio records
##
x$id <- ffseq_len(nrow(x))
xexploded <- expand.ffgrid(x$id, ff(1:100)) # Had to wait 3 minutes on my computer
colnames(xexploded) <- c("id","explosion.nr")
xexploded <- merge(xexploded, x, by.x="id", by.y="id", all.x=TRUE, all.y=FALSE) ## this uses merge.ffdf, might take 30 minutes
dim(xexploded) ## hopsa, 280 Mio records and 13.5Gb created
sum(.rambytes[vmode(xexploded)]) * (nrow(xexploded) * 9.31322575 * 10^(-10))
## And build the linear model again on the whole dataset
mymodel <- bigglm(payment ~ sex + age + place.served, data = xexploded)
summary(mymodel)
- That wasn't that easy or was it. Now your turn