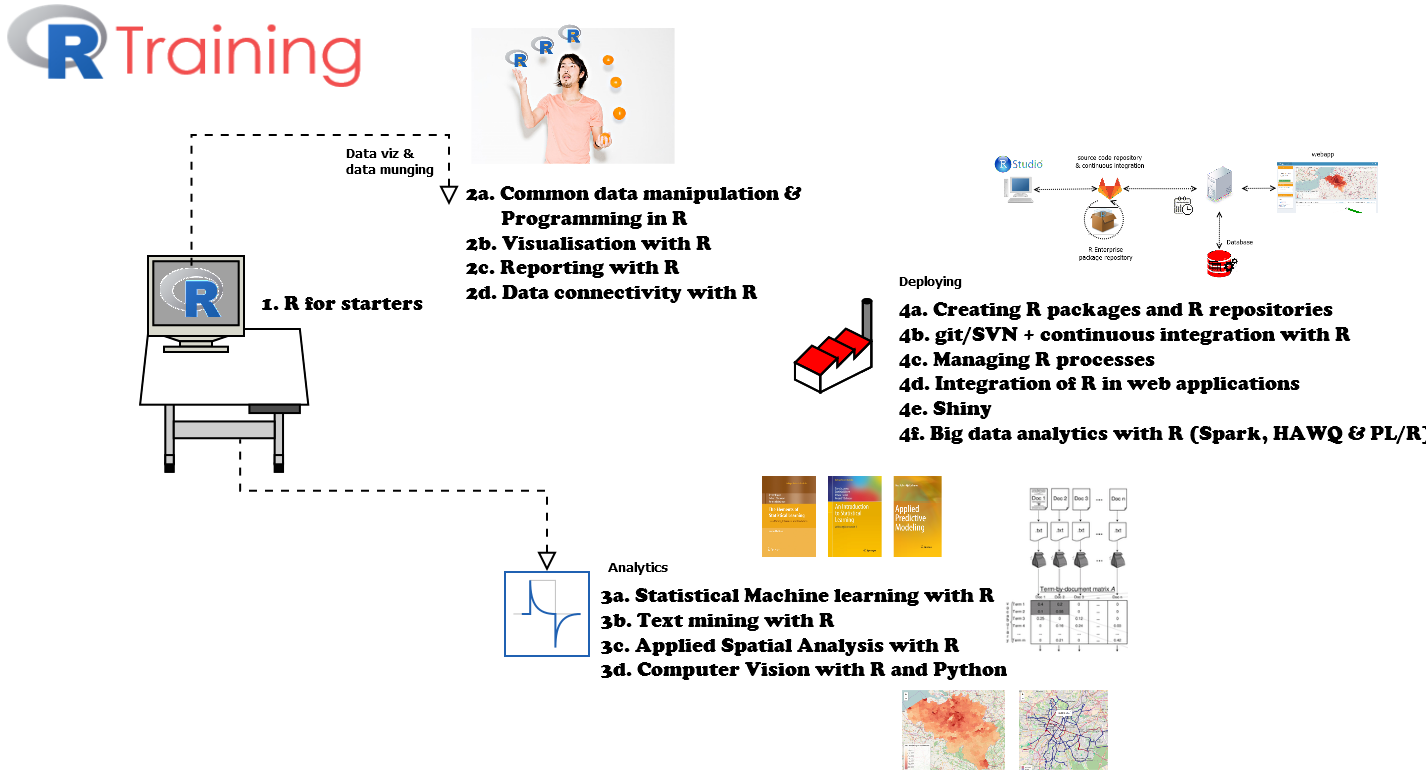
A few weeks ago, we pushed R package textplot to CRAN and it was accepted for release last week. The package contains straightforward functionalities for the visualisation of text, namely of
- text cooccurences
- text clusters (in casu biterm clusters)
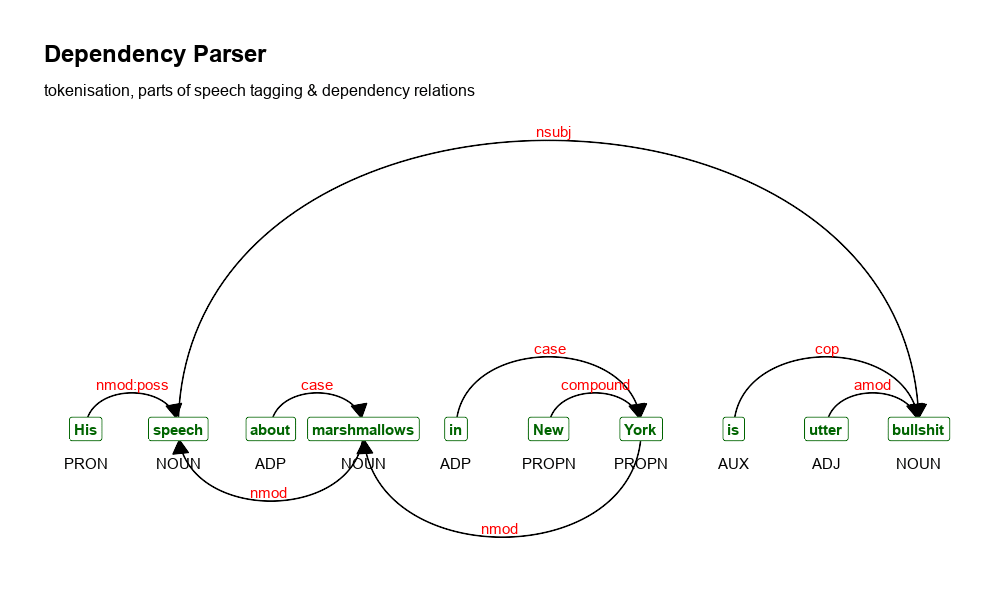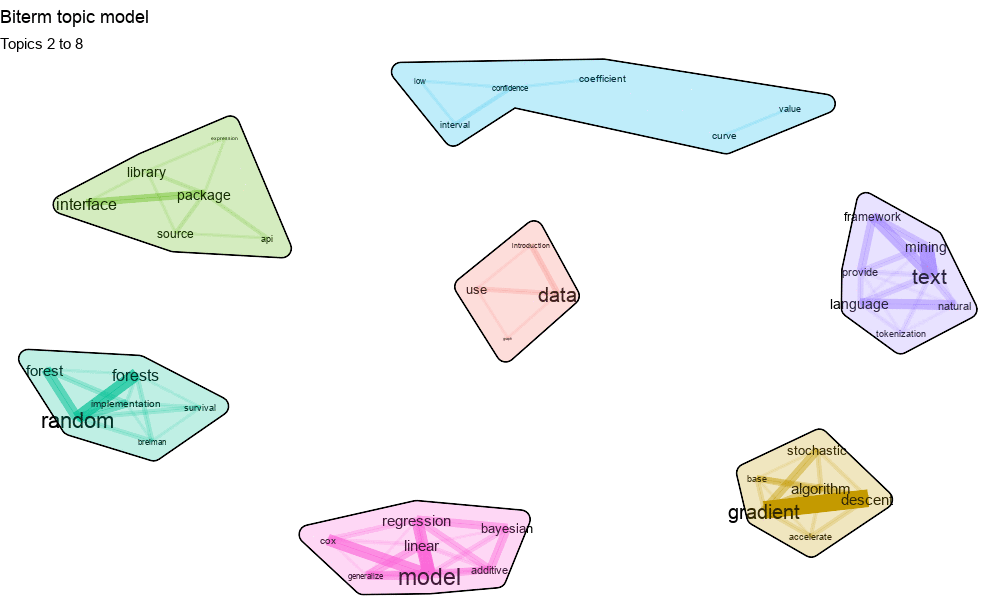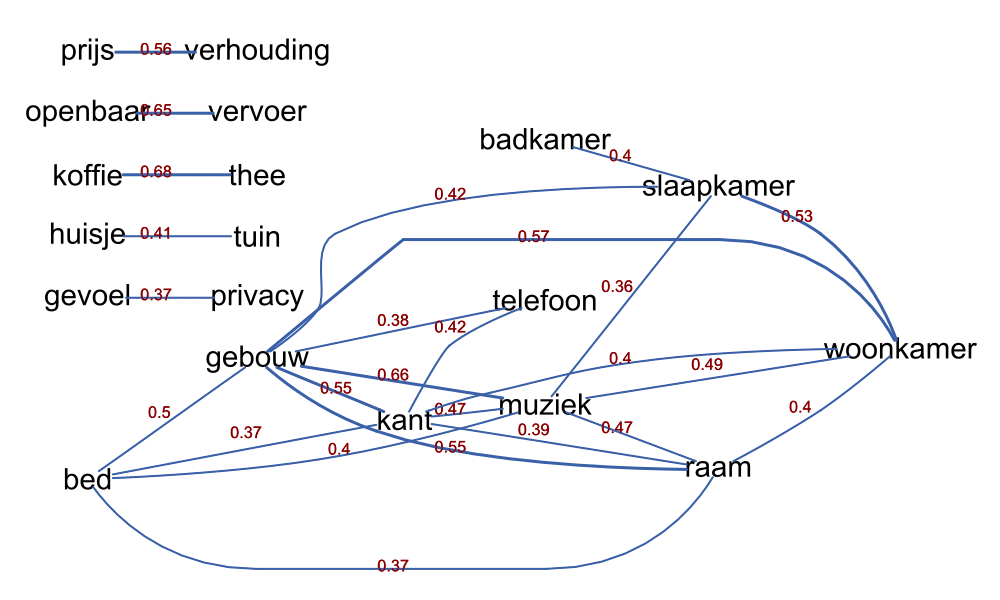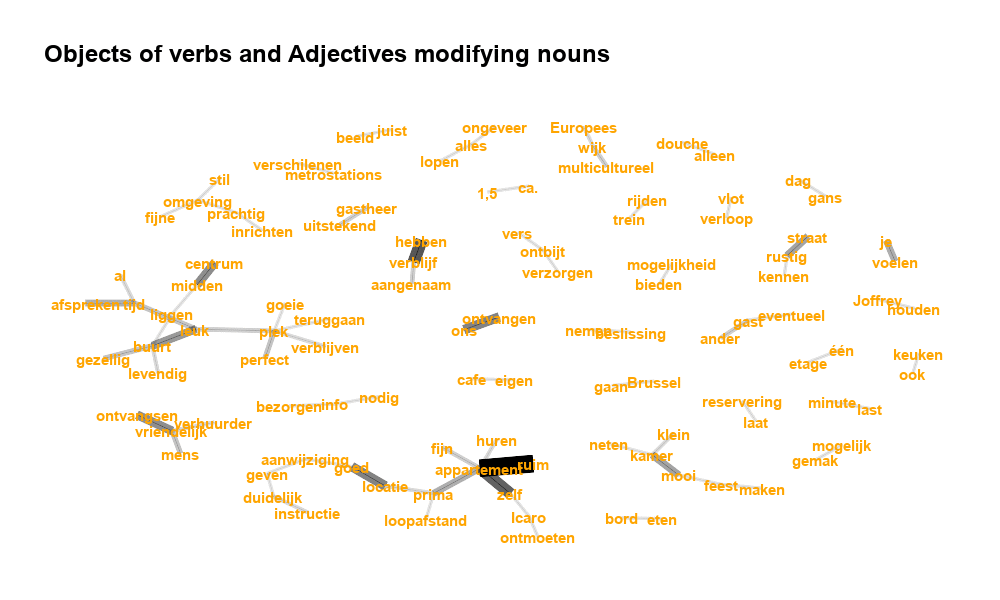
- dependency parsing results
- text correlations and text frequencies
Some examples of these plots are shown in the gif.
More details can be found in the pdf presentation shown below.
Enjoy.
A few weeks ago, we published an update of the BTM (Biterm Topic Models for text) package on CRAN.
Biterm Topic Models are especially usefull if you want to find topics in collections of short texts. Short texts are typically a twitter message, a short answer on a survey, the title of an email, search questions, ... . For these types of short texts traditional topic models like Latent Dirichlet Allocation are less suited as most information is available in short word combinations. The R package BTM finds topics in such short texts by explicitely modelling word-word co-occurrences (biterms) in a short window.
The update which was pushed to CRAN a few weeks ago now allows to explicitely provide a set of biterms to cluster upon. Let us show an example on clustering a subset of R package descriptions on CRAN. The resulting cluster visualisation looks like this.
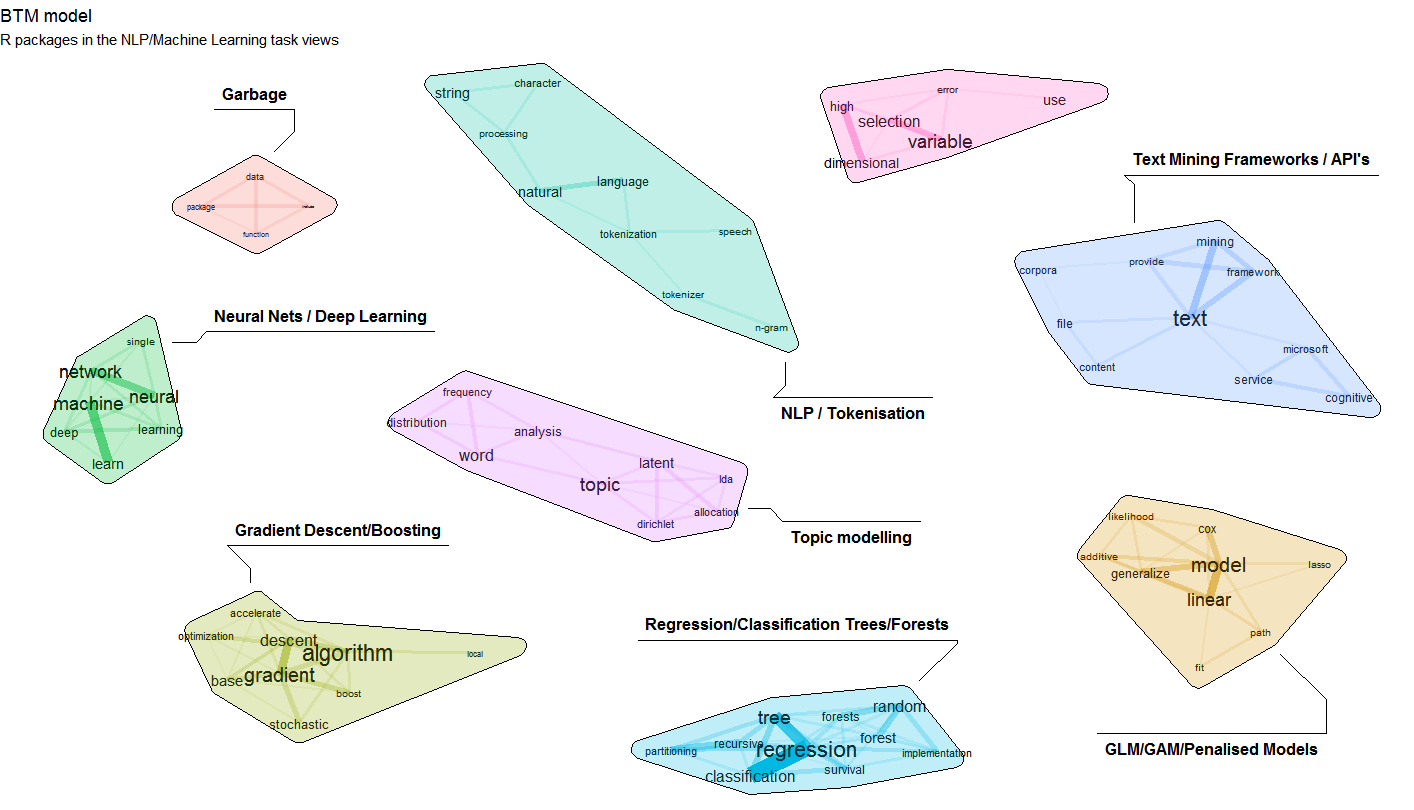
If you want to reproduce this, the following snippets show how to do this. Steps are as follows
1. Get some data of R packages and their description in plain text
## Get list of packages in the NLP/Machine Learning Task Views
library(ctv)
pkgs <- available.views()
names(pkgs) <- sapply(pkgs, FUN=function(x) x$name)
pkgs <- c(pkgs$NaturalLanguageProcessing$packagelist$name, pkgs$MachineLearning$packagelist$name)
## Get package descriptions of these packages
library(tools)
x <- CRAN_package_db()
x <- x[, c("Package", "Title", "Description")]
x$doc_id <- x$Package
x$text <- tolower(paste(x$Title, x$Description, sep = "\n"))
x$text <- gsub("'", "", x$text)
x$text <- gsub("<.+>", "", x$text)
x <- subset(x, Package %in% pkgs)
2. Use the udpipe R package to perform Parts of Speech tagging on the package title and descriptions and use udpipe as well for extracting cooccurrences of nouns, adjectives and verbs within 3 words distance.
library(udpipe)
library(data.table)
library(stopwords)
anno <- udpipe(x, "english", trace = 10)
biterms <- as.data.table(anno)
biterms <- biterms[, cooccurrence(x = lemma,
relevant = upos %in% c("NOUN", "ADJ", "VERB") &
nchar(lemma) > 2 & !lemma %in% stopwords("en"),
skipgram = 3),
by = list(doc_id)]
3. Build the biterm topic model with 9 topics and provide the set of biterms to cluster upon
library(BTM)
set.seed(123456)
traindata <- subset(anno, upos %in% c("NOUN", "ADJ", "VERB") & !lemma %in% stopwords("en") & nchar(lemma) > 2)
traindata <- traindata[, c("doc_id", "lemma")]
model <- BTM(traindata, biterms = biterms, k = 9, iter = 2000, background = TRUE, trace = 100)
4. Visualise the biterm topic clusters using the textplot package available at https://github.com/bnosac/textplot. This creates the plot show above.
library(textplot)
library(ggraph)
plot(model, top_n = 10,
title = "BTM model", subtitle = "R packages in the NLP/Machine Learning task views",
labels = c("Garbage", "Neural Nets / Deep Learning", "Topic modelling",
"Regression/Classification Trees/Forests", "Gradient Descent/Boosting",
"GLM/GAM/Penalised Models", "NLP / Tokenisation",
"Text Mining Frameworks / API's", "Variable Selection in High Dimensions"))
Enjoy!
I lost a few hours this afternoon when digging into the Corona virus data mainly caused by reading this article at this website which gives a nice view on how to be aware of potential issues which can arise when collecting data and to be aware of hidden factors and it also shows Belgium.
- As a Belgian, I was interested to see how Corona might impact our lives in the next weeks and out of curiosity I was interested to see how we are doing compared to other countries regarding containment of the Corona virus outspread - especially since we still do not have a government in Belgium after elections 1 year ago.
- In what follows, I'll be showing some graphs using data available at https://github.com/CSSEGISandData/COVID-19 (it provides up-to-date statistics on Corona cases). If you want to reproduce this, pull the repository and just execute the following R code shown.
Data
Let's see first if the data is exactly what is shown at our National Television.
library(data.table)
library(lattice)
x <- list.files("csse_covid_19_data/csse_covid_19_daily_reports/", pattern = ".csv", full.names = TRUE)
x <- data.frame(file = x, date = substr(basename(x), 1, 10), stringsAsFactors = FALSE)
x <- split(x$file, x$date)
x <- lapply(x, fread)
x <- rbindlist(x, fill = TRUE, idcol = "date")
x$date <- as.Date(x$date, format = "%m-%d-%Y")
x <- setnames(x,
old = c("date", "Country/Region", "Province/State", "Confirmed", "Deaths", "Recovered"),
new = c("date", "region", "subregion", "confirmed", "death", "recovered"))
x <- subset(x, subregion %in% "Hubei" |
region %in% c("Belgium", "France", "Netherlands", "Spain", "Singapore", "Germany", "Switzerland", "Italy"))
x$area <- ifelse(x$subregion %in% "Hubei", x$subregion, x$region)
x <- x[!duplicated(x, by = c("date", "area")), ]
x <- x[, c("date", "area", "confirmed", "death", "recovered")]
subset(x, area %in% "Belgium" & confirmed > 1)
Yes, the data from https://github.com/CSSEGISandData/COVID-19 looks correct indeed. Same numbers as reported on the Belgian Television.
| date | area | confirmed | death | recovered |
|---|
| 2020-03-01 |
Belgium |
2 |
0 |
1 |
| 2020-03-02 |
Belgium |
8 |
0 |
1 |
| 2020-03-03 |
Belgium |
13 |
0 |
1 |
| 2020-03-04 |
Belgium |
23 |
0 |
1 |
| 2020-03-05 |
Belgium |
50 |
0 |
1 |
| 2020-03-06 |
Belgium |
109 |
0 |
1 |
| 2020-03-07 |
Belgium |
169 |
0 |
1 |
| 2020-03-08 |
Belgium |
200 |
0 |
1 |
| 2020-03-09 |
Belgium |
239 |
0 |
1 |
| 2020-03-10 |
Belgium |
267 |
0 |
1 |
| 2020-03-11 |
Belgium |
314 |
3 |
1 |
Exponential number of cases of Corona
- Now is the outbreak really exponential? Let's make some graphs.
What is clear when looking at the plots is that indeed infections happen at a exponential scale except in Singapore where the government managed to completely isolate the Corona cases, while in Belgium and other European countries the government lacked the opportunity to isolate the Corona cases and we are now in a phase of trying to slow down to reduce and spread the impact.
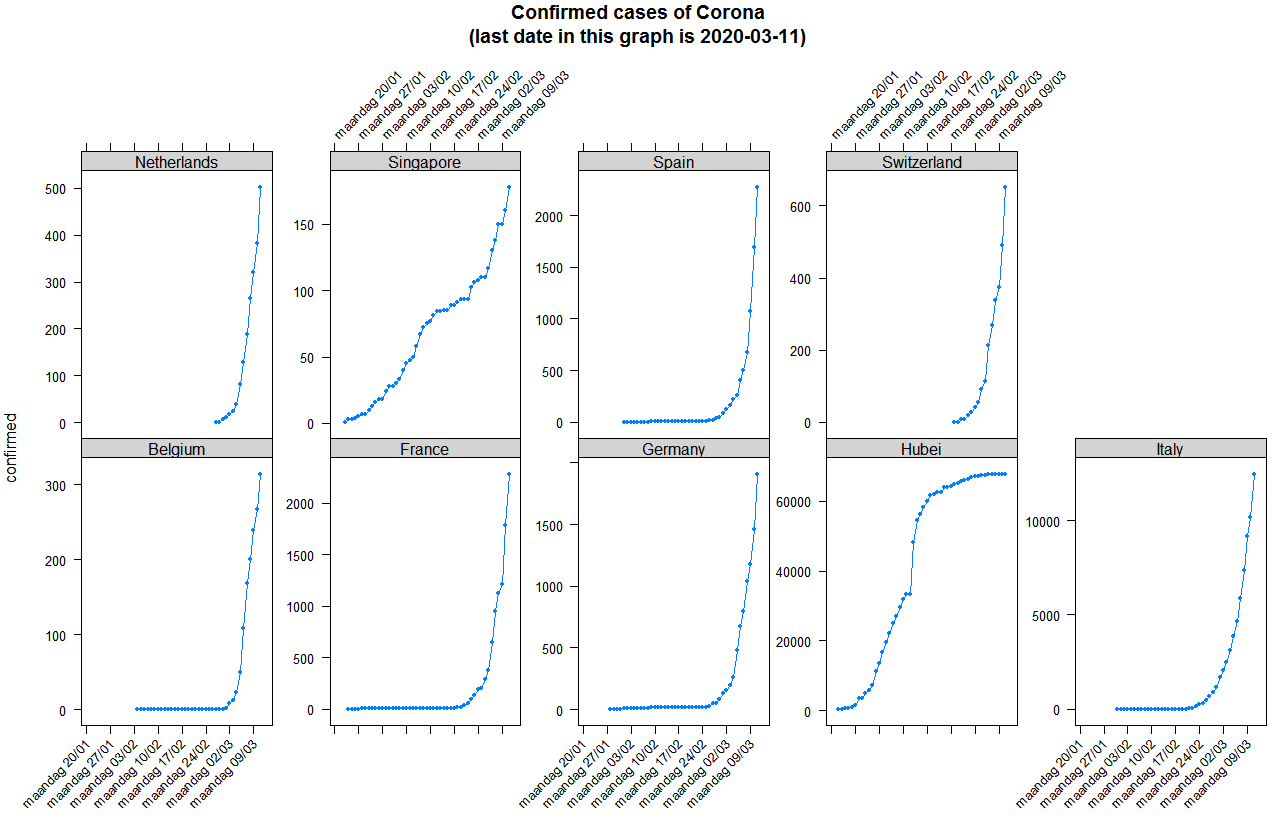
You can reproduce the plot as follows
trellis.par.set(strip.background = list(col = "lightgrey"))
xyplot(confirmed ~ date | area, data = x, type = "b", pch = 20,
scales = list(y = list(relation = "free", rot = 0), x = list(rot = 45, format = "%A %d/%m")),
layout = c(5, 2), main = sprintf("Confirmed cases of Corona\n(last date in this graph is %s)", max(x$date)))
Compare to other countries - onset
It is clear that the onset of Corona is different in each country. Let's define the onset (day 0) as the day where 75 persons had Corona in the country. That will allow us to compare different countries. In Belgium we started to have more than 75 patients with Corona on Friday 2020-03-06. In the Netherlands that was one day earlier.
| date | area | confirmed |
|---|
| 2020-01-22 |
Hubei |
444 |
| 2020-02-17 |
Singapore |
77 |
| 2020-02-23 |
Italy |
155 |
| 2020-02-29 |
Germany |
79 |
| 2020-02-29 |
France |
100 |
| 2020-03-01 |
Spain |
84 |
| 2020-03-04 |
Switzerland |
90 |
| 2020-03-05 |
Netherlands |
82 |
| 2020-03-06 |
Belgium |
109 |
Reproduce as follows:
x <- x[order(x$date, x$area, decreasing = TRUE), ]
x <- x[, days_since_case_onset := as.integer(date - min(date[confirmed > 75])), by = list(area)]
x <- x[, newly_confirmed := as.integer(confirmed - shift(confirmed, n = 1, type = "lead")), by = list(area)]
onset <- subset(x, days_since_case_onset == 0, select = c("date", "area", "confirmed"))
onset[order(onset$date), ]
Compare to other countries - what can we expect?
- Now are we doing better than other countries in the EU?
Following plot shows the log of the number of people diagnosed as having Corona since the onset date shown above. It looks like Belgium has learned a bit from the issues in Italy but it still hasn't learned the way to deal with the virus outbreak the same as e.g. Singapore has done (a country which learned from the SARS outbreak).
Based on the blue line, we can expect Belgium to have next week between roughly 1100 confirmed cases (log(1100)=7) or if we follow the trend of France that would be roughly 3000 (log(3000)=8) patients with Corona. We hope that it is only the first.
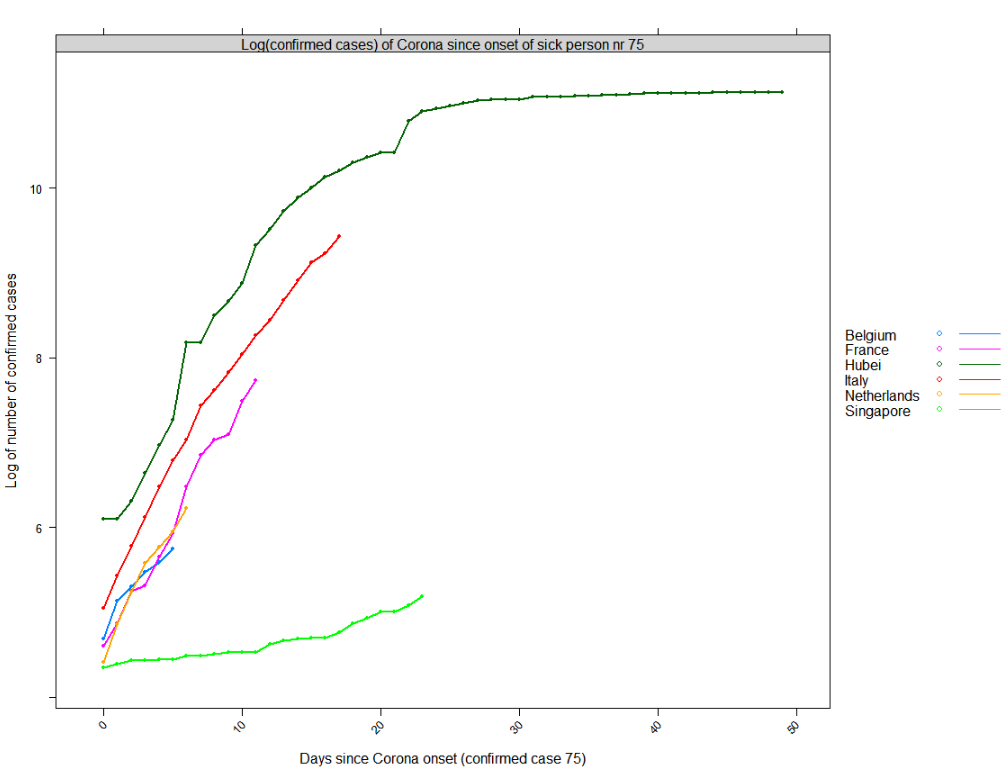
Reproduce as follows:
xyplot(log(confirmed) ~ days_since_case_onset | "Log(confirmed cases) of Corona since onset of sick person nr 75",
groups = area,
data = subset(x, days_since_case_onset >= 0 &
area %in% c("Hubei", "France", "Belgium", "Singapore", "Netherlands", "Italy")),
xlab = "Days since Corona onset (confirmed case 75)", ylab = "Log of number of confirmed cases",
auto.key = list(space = "right", lines = TRUE),
type = "b", pch = 20, lwd = 2)
Compared to the Netherlands
- Now, are we doing better than The Netherlands?
Currently it looks like we are. But time will tell. Given the trend shown above, I can only hope everyone in Belgium follows the government guidelines as strict as possible.
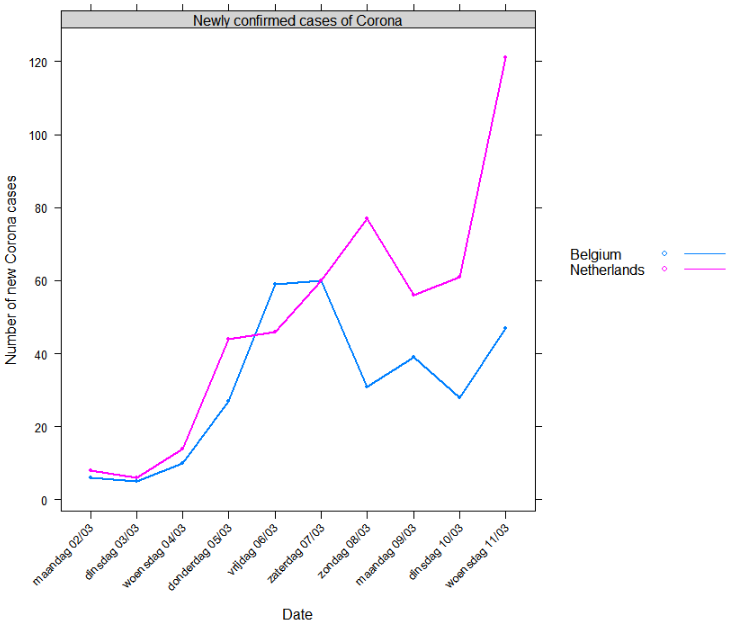
Reproduce as follows:
xyplot(newly_confirmed ~ date | "Newly confirmed cases of Corona", groups = area,
data = subset(x, area %in% c("Belgium", "Netherlands") & date > as.Date("2020-03-01")),
xlab = "Date", ylab = "Number of new Corona cases",
scales = list(x = list(rot = 45, format = "%A %d/%m", at = seq(as.Date("2020-03-01"), Sys.Date(), by = "day"))),
auto.key = list(space = "right", lines = TRUE),
type = "b", pch = 20, lwd = 2)
 You can view the presentation below.
You can view the presentation below.